| Raw reads | .
|
| Replicate 1 | .
|
| Fastq (41M) | /mnt/storage/home/psbelokopytova/CNTN6_new_father/fastq/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.fastq.gz
|
| Replicate 2 | .
|
| Fastq (44M) | /mnt/storage/home/psbelokopytova/CNTN6_new_father/fastq/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.fastq.gz
|
| Control 1 | .
|
| Fastq (41M) | /mnt/storage/home/psbelokopytova/CNTN6_new_father/fastq/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.fastq.gz
|
| Alignment | .
|
| Replicate 1 | .
|
| Bam (40M) | ./align/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.bam
|
| Filtered & deduped bam (29M) | ./align/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.bam
|
| Tag-align | ./align/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.tagAlign.gz
|
| Replicate 2 | .
|
| Bam (43M) | ./align/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.bam
|
| Filtered & deduped bam (32M) | ./align/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.bam
|
| Tag-align | ./align/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.tagAlign.gz
|
| Control 1 | .
|
| Bam (40M) | ./align/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.bam
|
| Filtered & deduped bam (32M) | ./align/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.bam
|
| Tag-align | ./align/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.gz
|
| Pooled replicate | .
|
| Tag-align | ./align/pooled_rep/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup_pooled.tagAlign.gz
|
| Pseudo-replicates | .
|
| Replicate 1 | .
|
| Pseudo-replicate 1 | .
|
| Tag-align | ./align/pseudo_reps/rep1/pr1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr1.tagAlign.gz
|
| Pseudo-replicate 2 | .
|
| Tag-align | ./align/pseudo_reps/rep1/pr2/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr2.tagAlign.gz
|
| Replicate 2 | .
|
| Pseudo-replicate 1 | .
|
| Tag-align | ./align/pseudo_reps/rep2/pr1/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.pr1.tagAlign.gz
|
| Pseudo-replicate 2 | .
|
| Tag-align | ./align/pseudo_reps/rep2/pr2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.pr2.tagAlign.gz
|
| Pooled pseudo-replicates | .
|
| Pooled pseudo-replicate 1 | .
|
| Tag-align | ./align/pooled_pseudo_reps/ppr1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr1_pooled.tagAlign.gz
|
| Pooled pseudo-replicate 2 | .
|
| Tag-align | ./align/pooled_pseudo_reps/ppr2/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr2_pooled.tagAlign.gz
|
| Signal tracks | .
|
| MACS2 | .
|
| Replicate 1 | .
|
| P-value | ./signal/macs2/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval.signal.bw
|
| Fold enrichment | ./signal/macs2/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.fc.signal.bw
|
| Replicate 2 | .
|
| P-value | ./signal/macs2/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval.signal.bw
|
| Fold enrichment | ./signal/macs2/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.fc.signal.bw
|
| Pooled replicate | .
|
| P-value | ./signal/macs2/pooled_rep/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup_pooled.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval.signal.bw
|
| Fold enrichment | ./signal/macs2/pooled_rep/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup_pooled.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.fc.signal.bw
|
| Peaks | .
|
| MACS2 | .
|
| Replicate 1 | .
|
| Narrow peak | ./peak/macs2/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Replicate 2 | .
|
| Narrow peak | ./peak/macs2/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pooled replicate | .
|
| Narrow peak | ./peak/macs2/pooled_rep/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup_pooled.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pseudo-replicates | .
|
| Replicate 1 | .
|
| Pseudo-replicate 1 | .
|
| Narrow peak | ./peak/macs2/pseudo_reps/rep1/pr1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr1.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pseudo-replicate 2 | .
|
| Narrow peak | ./peak/macs2/pseudo_reps/rep1/pr2/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr2.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Replicate 2 | .
|
| Pseudo-replicate 1 | .
|
| Narrow peak | ./peak/macs2/pseudo_reps/rep2/pr1/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.pr1.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pseudo-replicate 2 | .
|
| Narrow peak | ./peak/macs2/pseudo_reps/rep2/pr2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.pr2.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pooled pseudo-replicate | .
|
| Pooled pseudo-replicate 1 | .
|
| Narrow peak | ./peak/macs2/pooled_pseudo_reps/ppr1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr1_pooled.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Pooled pseudo-replicate 2 | .
|
| Narrow peak | ./peak/macs2/pooled_pseudo_reps/ppr2/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pr2_pooled.tagAlign_x_P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.tagAlign.pval0.01.500K.narrowPeak.gz
|
| Naive overlap | .
|
| True replicates | .
|
| Rep. 1 vs. Rep. 2 | .
|
| Overlapping peak | ./peak/macs2/overlap/true_reps/rep1-rep2/14-10_K27_rep1-rep2.naive_overlap.filt.narrowPeak.gz
|
| Pseudo-replicates | .
|
| Replicate 1 | .
|
| Overlapping peak | ./peak/macs2/overlap/pseudo_reps/rep1/14-10_K27_rep1-pr.naive_overlap.filt.narrowPeak.gz
|
| Replicate 2 | .
|
| Overlapping peak | ./peak/macs2/overlap/pseudo_reps/rep2/14-10_K27_rep2-pr.naive_overlap.filt.narrowPeak.gz
|
| Pooled pseudo-replicates | .
|
| Overlapping peak | ./peak/macs2/overlap/pooled_pseudo_reps/14-10_K27_ppr.naive_overlap.filt.narrowPeak.gz
|
| Optimal set | .
|
| Overlapping peak | ./peak/macs2/overlap/optimal_set/14-10_K27_ppr.naive_overlap.filt.narrowPeak.gz
|
| Conservative set | .
|
| Overlapping peak | ./peak/macs2/overlap/conservative_set/14-10_K27_rep1-rep2.naive_overlap.filt.narrowPeak.gz
|
| QC and logs | .
|
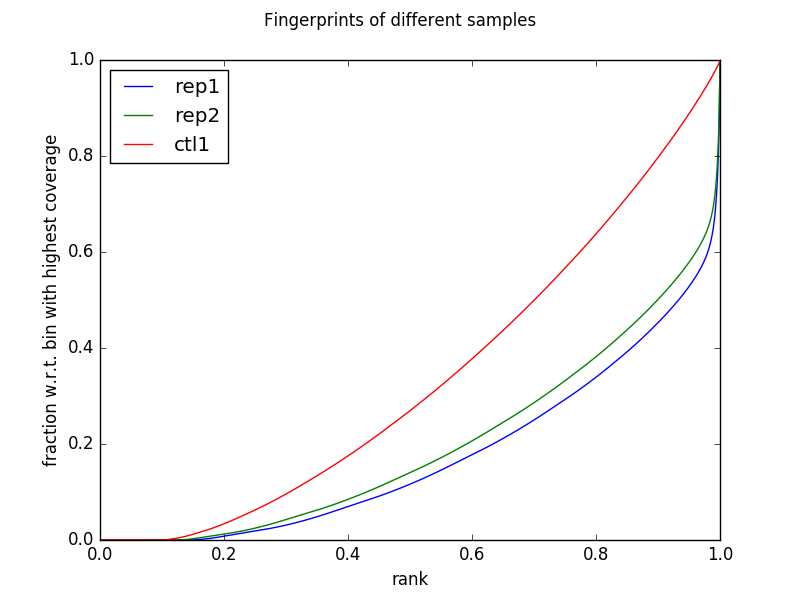
| Fingerprint JSD Plot | ./qc/14-10_K27_jsd.png
|
| Replicate 1 | .
|
| Filtered flagstat log | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.flagstat.qc
|
| JS Distance | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.jsd.qc
|
| BWA map. flagstat log | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.flagstat.qc
|
| Dedup. log | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.dup.qc
|
| PBC log | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.nodup.pbc.qc
|
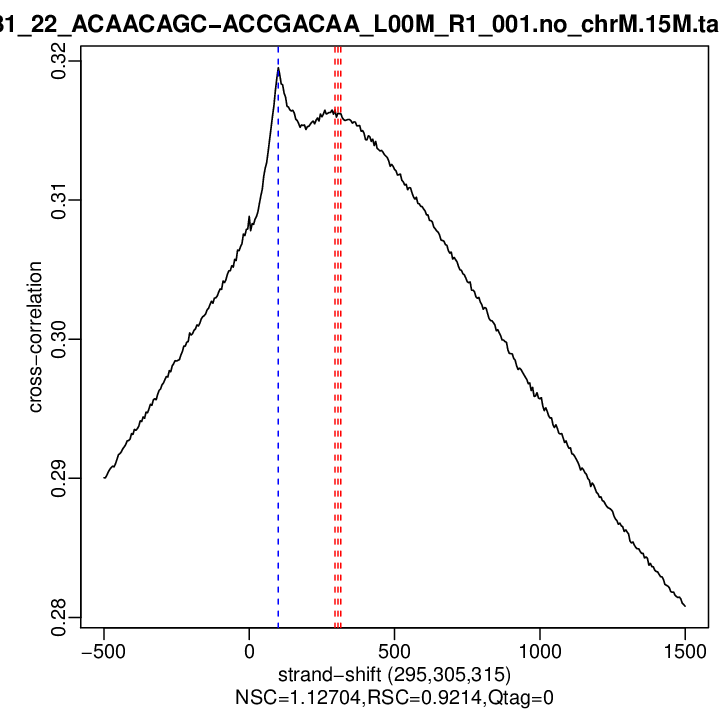
| Cross-corr. log | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.no_chrM.15M.cc.qc
|
| Cross-corr. plot | ./qc/rep1/P381_22_ACAACAGC-ACCGACAA_L00M_R1_001.no_chrM.15M.cc.plot.pdf
|
| Replicate 2 | .
|
| Filtered flagstat log | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.flagstat.qc
|
| JS Distance | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.jsd.qc
|
| BWA map. flagstat log | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.flagstat.qc
|
| Dedup. log | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.dup.qc
|
| PBC log | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.nodup.pbc.qc
|
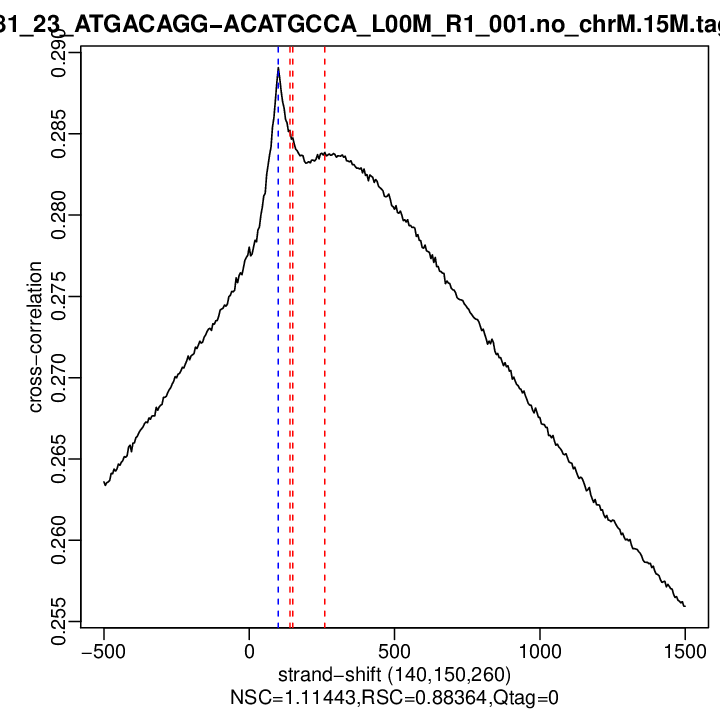
| Cross-corr. log | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.no_chrM.15M.cc.qc
|
| Cross-corr. plot | ./qc/rep2/P381_23_ATGACAGG-ACATGCCA_L00M_R1_001.no_chrM.15M.cc.plot.pdf
|
| Control 1 | .
|
| Filtered flagstat log | ./qc/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.flagstat.qc
|
| BWA map. flagstat log | ./qc/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.flagstat.qc
|
| Dedup. log | ./qc/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.dup.qc
|
| PBC log | ./qc/ctl1/P381_7_CTAGCTCA-AACTGAGG_L00M_R1_001.nodup.pbc.qc
|
| Naive overlap | .
|
| Overlap QC log | ./qc/14-10_K27_peak_overlap_final.qc
|
| True replicates | .
|
| Rep. 1 vs. Rep. 2 | .
|
| FRiP | ./peak/macs2/overlap/true_reps/rep1-rep2/14-10_K27_rep1-rep2.naive_overlap.filt.narrowPeak.FRiP.qc
|
| Pseudo-replicates | .
|
| Replicate 1 | .
|
| FRiP | ./peak/macs2/overlap/pseudo_reps/rep1/14-10_K27_rep1-pr.naive_overlap.filt.narrowPeak.FRiP.qc
|
| Replicate 2 | .
|
| FRiP | ./peak/macs2/overlap/pseudo_reps/rep2/14-10_K27_rep2-pr.naive_overlap.filt.narrowPeak.FRiP.qc
|